Learning Tumor Evolution with Diffusion Models
Jul 28, 2025
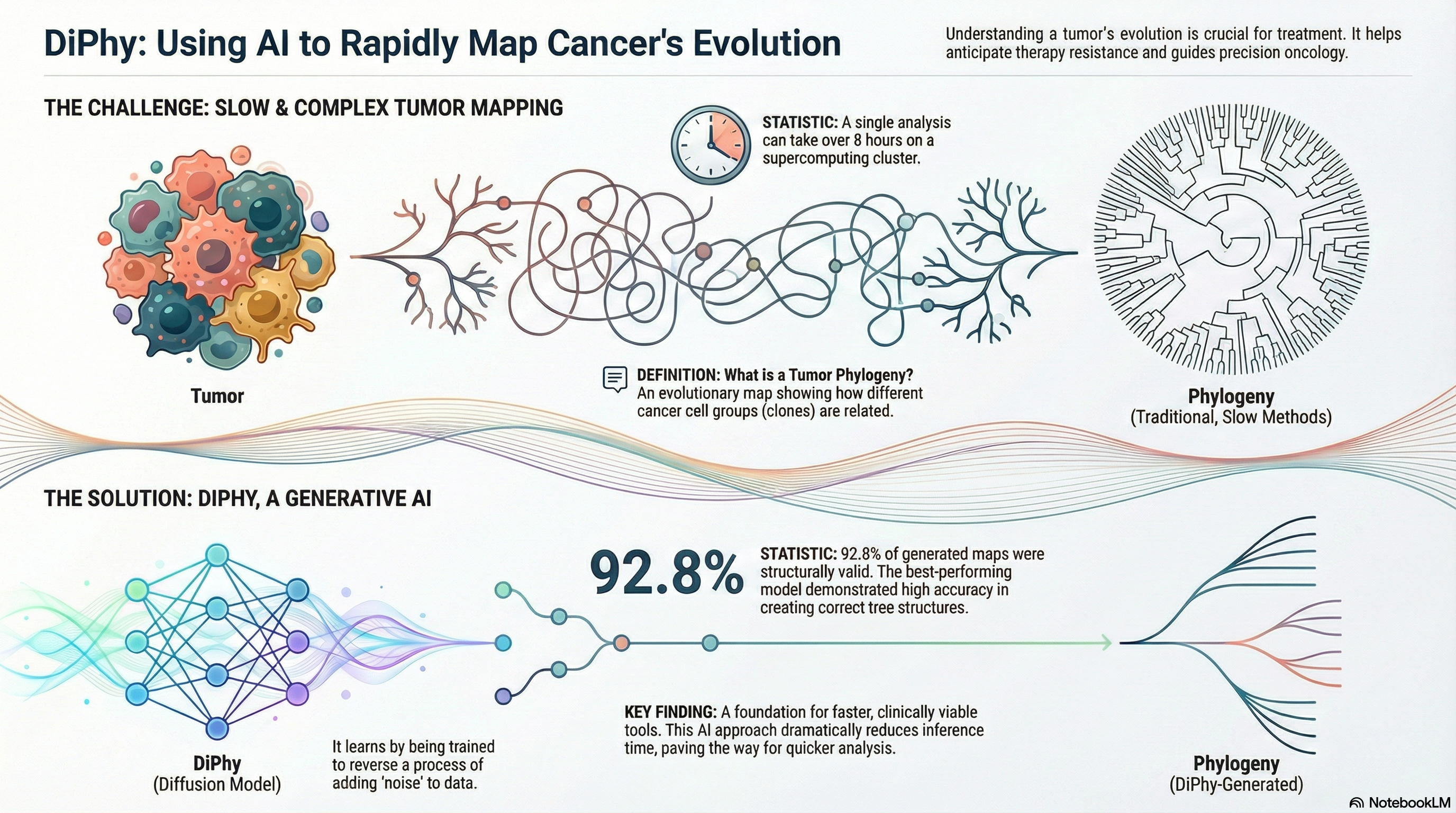
Reconstructing tumor clonal phylogenies is vital for understanding intra-tumor heterogeneity, anticipating therapy resistance, and advancing precision oncology. Traditional unsupervised model-based approaches infer evolutionary trees from bulk sequencing data but are constrained by lengthy runtimes and instability when applied to sparse datasets. Data-driven generative modeling offers an alternative by directly learning complex statistical distributions. Learning distributions of tumor phylogenies enables greater generalizability and faster inference.
This work explores the mathematical feasibility of using generative discrete diffusion for phylogenetic reconstruction. We introduce DiPhy, an adapted discrete graph diffusion model for unconditional phylogeny generation. We develop a synthetic training dataset and train two 7.1 million parameter models on purely synthetic data, achieving 92.4% structurally valid graphs at test time. This work serves as an initial step toward leveraging generative deep learning for fast, reliable tumor phylogeny inference.